Clustering of movement-model parameters
cluster.RdThese functions cluster and classify individual movement models and related estimates, including AKDE home-range areas, while taking into account estimation uncertainty.
cluster(x,level=0.95,level.UD=0.95,debias=TRUE,IC="BIC",units=TRUE,plot=TRUE,sort=FALSE,
...)Arguments
- x
A list of
ctmmmovement-model objects,UDobjects, orUDsummaryoutput, constituting a sampled population, or a list of such lists, each constituting a sampled sub-population.- level
Confidence level for parameter estimates.
- level.UD
Coverage level for home-range estimates. E.g., 50% core home range.
- debias
Apply Bessel's inverse-Gaussian correction and various other bias corrections.
- IC
Information criterion to determine whether or not population variation can be estimated. Can be
"AICc",AIC, or"BIC".- units
Convert result to natural units.
- plot
Generate a meta-analysis forest plot with two means.
- sort
Sort individuals by their point estimates in forest plot.
- ...
Further arguments passed to
plot.
Details
So-far only the clustering of home-range areas is implemented. More details will be provided in an upcomming manuscript.
Value
A list with elements P and CI,
where P is an array of individual membership probabilities for sub-population 1,
and CI is a table with rows corresponding to the sub-population means, coefficients of variation, and membership probabilities, and the ratio of sub-population means.
Note
The AICc formula is approximated via the Gaussian relation.
Examples
# \donttest{
# load package and data
library(ctmm)
data(buffalo)
# fit movement models
FITS <- AKDES <- list()
for(i in 1:length(buffalo))
{
GUESS <- ctmm.guess(buffalo[[i]],interactive=FALSE)
# use ctmm.select unless you are certain that the selected model is OUF
FITS[[i]] <- ctmm.fit(buffalo[[i]],GUESS)
}
# calculate AKDES on a consistent grid
AKDES <- akde(buffalo,FITS)
#> Default grid size of 3 minutes chosen for bandwidth(...,fast=TRUE).
#> Default grid size of 2 minutes chosen for bandwidth(...,fast=TRUE).
#> Default grid size of 2 minutes chosen for bandwidth(...,fast=TRUE).
#> Default grid size of 3 minutes chosen for bandwidth(...,fast=TRUE).
#> Default grid size of 2 minutes chosen for bandwidth(...,fast=TRUE).
#> Default grid size of 5 minutes chosen for bandwidth(...,fast=TRUE).
# color to be spatially distinct
COL <- color(AKDES,by='individual')
# plot AKDEs
plot(AKDES,col.DF=COL,col.level=COL,col.grid=NA,level=NA)
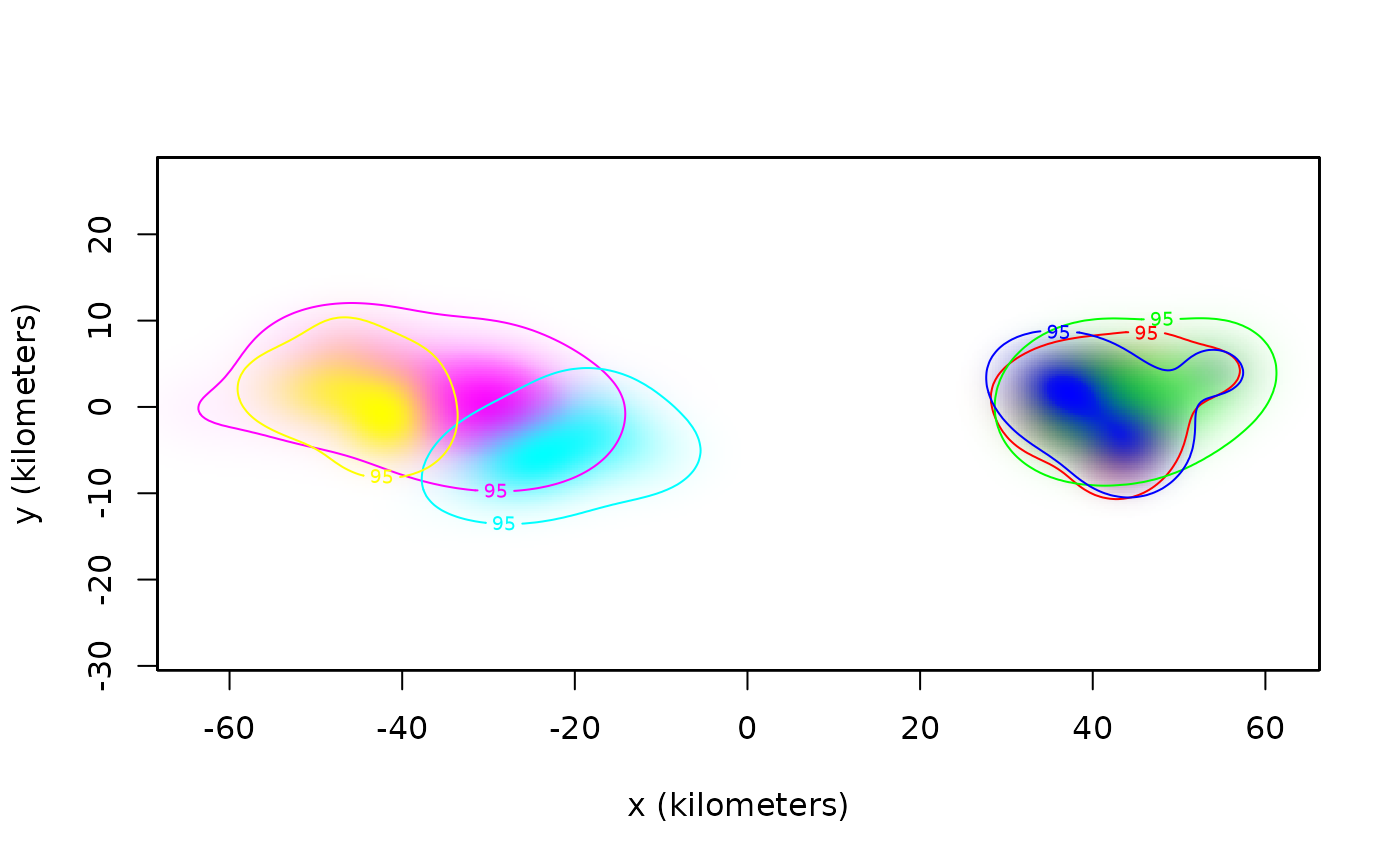 # cluster-analysis of buffalo
cluster(AKDES,sort=TRUE)
#> ΔBIC
#> Dirac-δ 0.0000000
#> inverse-Gaussian 0.9591028
#> Dirac-δ + Dirac-δ 1.3582120
#> inverse-Gaussian(CoV) + inverse-Gaussian(CoV) 3.1499715
#> Dirac-δ + inverse-Gaussian 3.1499715
#> inverse-Gaussian + Dirac-δ 3.1499715
#> inverse-Gaussian(CoV₁) + inverse-Gaussian(CoV₂) 4.9417310
# cluster-analysis of buffalo
cluster(AKDES,sort=TRUE)
#> ΔBIC
#> Dirac-δ 0.0000000
#> inverse-Gaussian 0.9591028
#> Dirac-δ + Dirac-δ 1.3582120
#> inverse-Gaussian(CoV) + inverse-Gaussian(CoV) 3.1499715
#> Dirac-δ + inverse-Gaussian 3.1499715
#> inverse-Gaussian + Dirac-δ 3.1499715
#> inverse-Gaussian(CoV₁) + inverse-Gaussian(CoV₂) 4.9417310
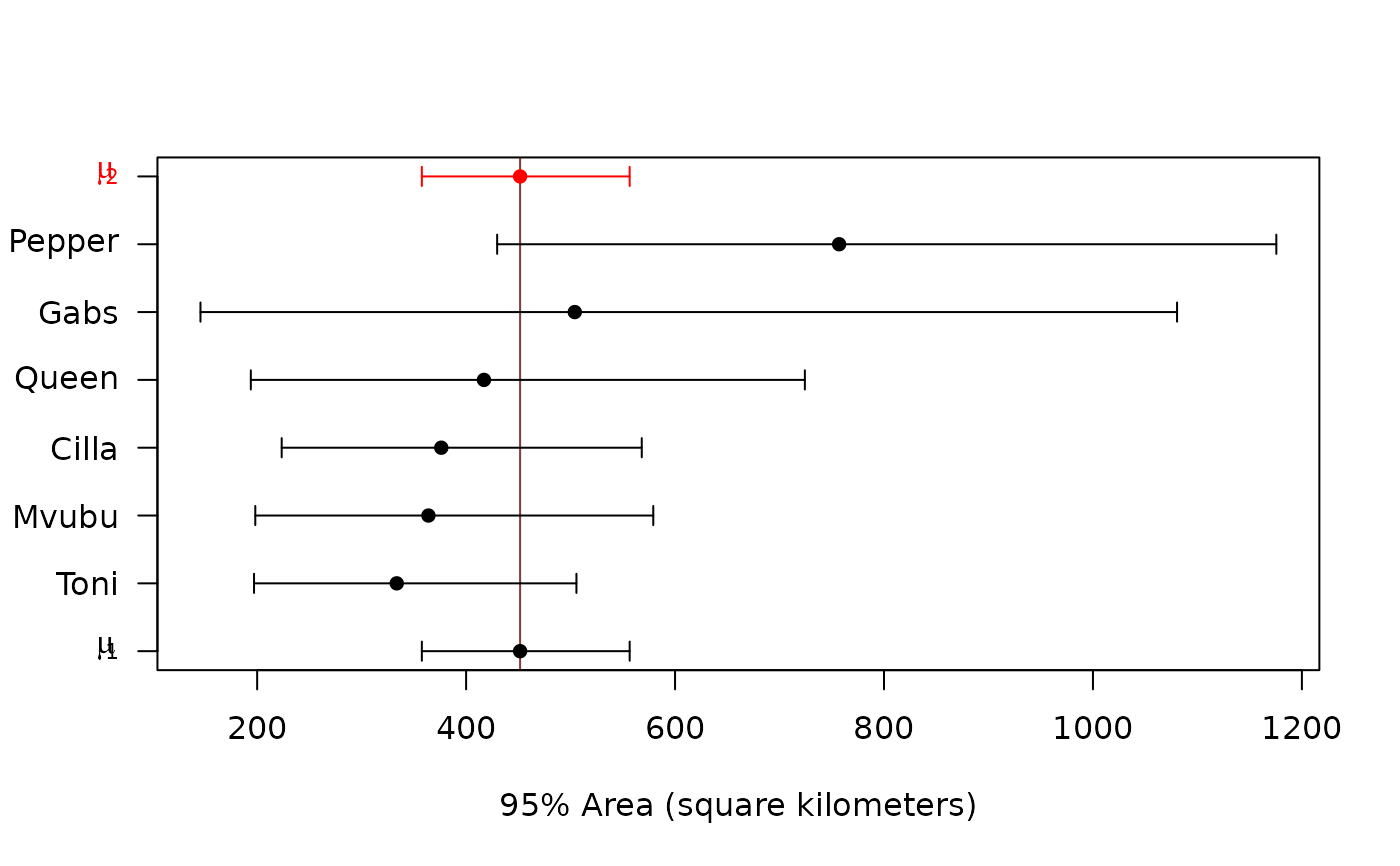 #> $P
#> Cilla Gabs Mvubu Pepper Queen Toni
#> 1 1 1 1 1 1
#>
#> $CI
#> low est high
#> μ₁ (km²) 357.629 451.6807 556.5461
#> CoV₁ 0.000 0.0000 Inf
#> μ₂ (km²) 357.629 451.6807 556.5461
#> CoV₂ 0.000 0.0000 Inf
#> P₁ 0.000 1.0000 1.0000
#> P₂ 0.000 0.0000 1.0000
#> μ₂/μ₁ 0.000 1.0000 Inf
#>
# }
#> $P
#> Cilla Gabs Mvubu Pepper Queen Toni
#> 1 1 1 1 1 1
#>
#> $CI
#> low est high
#> μ₁ (km²) 357.629 451.6807 556.5461
#> CoV₁ 0.000 0.0000 Inf
#> μ₂ (km²) 357.629 451.6807 556.5461
#> CoV₂ 0.000 0.0000 Inf
#> P₁ 0.000 1.0000 1.0000
#> P₂ 0.000 0.0000 1.0000
#> μ₂/μ₁ 0.000 1.0000 Inf
#>
# }