Calculate an autocorrelated kernel density estimate
akde.RdThese functions calculate individual and population-level autocorrelated kernel density home-range estimates from telemetry data and a corresponding continuous-time movement models.
akde(data,CTMM,VMM=NULL,R=list(),SP=NULL,SP.in=TRUE,variable="utilization",debias=TRUE,
weights=FALSE,smooth=TRUE,error=0.001,res=10,grid=NULL,...)
pkde(data,UD,kernel="individual",weights=FALSE,ref="Gaussian",...)Arguments
- data
2D timeseries telemetry data represented as a
telemetryobject or list of objects.- CTMM
A
ctmmmovement model from the output ofctmm.fitor list of objects.- VMM
An optional vertical
ctmmobject for 3D home-range calculation.
- R
A named list of raster covariates if
CTMMcontains an RSF model.- SP
SpatialPolygonsDataFrame object for enforcing hard boundaries.
- SP.in
Locations are assumed to be inside the
SPpolygons ifSP.in=TRUEand outside ofSPifSP.in=FALSE.- variable
Not yet supported.
- debias
Debias the distribution for area estimation (AKDEc).
- smooth
"Smooth" out errors from the data.
- weights
Optimally weight the data to account for sampling bias (See
bandwidthforakdedetails).- error
Target probability error.
- res
Number of grid points along each axis, relative to the bandwidth.
- grid
Optional grid specification via
raster,UD, or list of arguments (See `Details' below).- ...
- UD
A list of individual
UDobjects corresponding todata.- kernel
Bandwidths are proportional to the individual covariances if
kernel="individual"or to the population covariance ifkernel="population".- ref
Include non-Gaussian overlap corrections if
ref="AKDE"andweights=TRUE.
Details
For weighted AKDE, please note additional ... arguments passed to bandwidth, which can have a large impact on computation time in certain cases.
When feeding in lists of telemetry and ctmm objects, all UDs will be calculated on the same grid. These UDs can be averaged with the mean.UD command.
If a UD or raster object is supplied in the grid argument, then the estimate will be calculated on the same grid. Alternatively, a list of grid arguments can be supplied, with any of the following components:
rA list with vectors
xandythat define the grid-cell midpoints.drA vector setting the
xandycell widths in meters. Equivalent toresforrasterobjects.extentThe \(x\)-\(y\) extent of the grid cells, formatted as from the output of
extent.align.to.originLogical value indicating that cell midpoint locations are aligned to be an integer number of
drsteps from the projection origin.
Value
Returns a UD object: a list with the sampled grid line locations r$x and r$y, the extent of each grid cell dr, the probability density and cumulative distribution functions evaluated on the sampled grid locations PDF & CDF, the optimal bandwidth matrix H, and the effective sample size of the data in DOF.H.
References
C. H. Fleming, W. F. Fagan, T. Mueller, K. A. Olson, P. Leimgruber, J. M. Calabrese, ``Rigorous home-range estimation with movement data: A new autocorrelated kernel-density estimator'', Ecology, 96:5, 1182-1188 (2015) doi:10.1890/14-2010.1 .
C. H. Fleming, J. M. Calabrese, ``A new kernel-density estimator for accurate home-range and species-range area estimation'', Methods in Ecology and Evolution, 8:5, 571-579 (2017) doi:10.1111/2041-210X.12673 .
C. H. Fleming, D. Sheldon, W. F. Fagan, P. Leimgruber, T. Mueller, D. Nandintsetseg, M. J. Noonan, K. A. Olson, E. Setyawan, A. Sianipar, J. M. Calabrese, ``Correcting for missing and irregular data in home-range estimation'', Ecological Applications, 28:4, 1003-1010 (2018) doi:10.1002/eap.1704 .
Note
In the case of coarse grids, the value of PDF in a grid cell corresponds to the average probability density over the entire rectangular cell.
The PDF estimate is not re-normalized to 1, and may fall short of this by the target numerical error. If inspecting quantiles that are very far from the data, the quantiles may hit the grid boundary or become erratic, making it necessary to reduce the numerical error target. However, default arguments should be able to render any quantiles of reasonable accuracy.
Prior to ctmm v0.3.2, the default AKDE method was the autocorrelated Gaussian reference function bandwidth.
Starting in v0.3.2, the default AKDE method is the autocorrelated Gaussian reference function bandwidth with debiased area.
Prior to ctmm v0.3.1, AKDEs included only errors due to autocorrelation uncertainty, which are insignificant in cases such as IID data.
Starting in v0.3.1, akde calculated an effective sample size DOF.H and used this to estimate area uncertainty under a Gaussian reference function approxmation.
In v0.3.2, this method was further improved to use DOF.area from the Gaussian reference function approximation.
See also
Examples
# \donttest{
# Load package and data
library(ctmm)
data(buffalo)
DATA <- buffalo$Cilla
# calculate fit guess object
GUESS <- ctmm.guess(DATA,interactive=FALSE)
# in general, you should be running ctmm.select here instead of ctmm.fit
FIT <- ctmm.fit(DATA,GUESS)
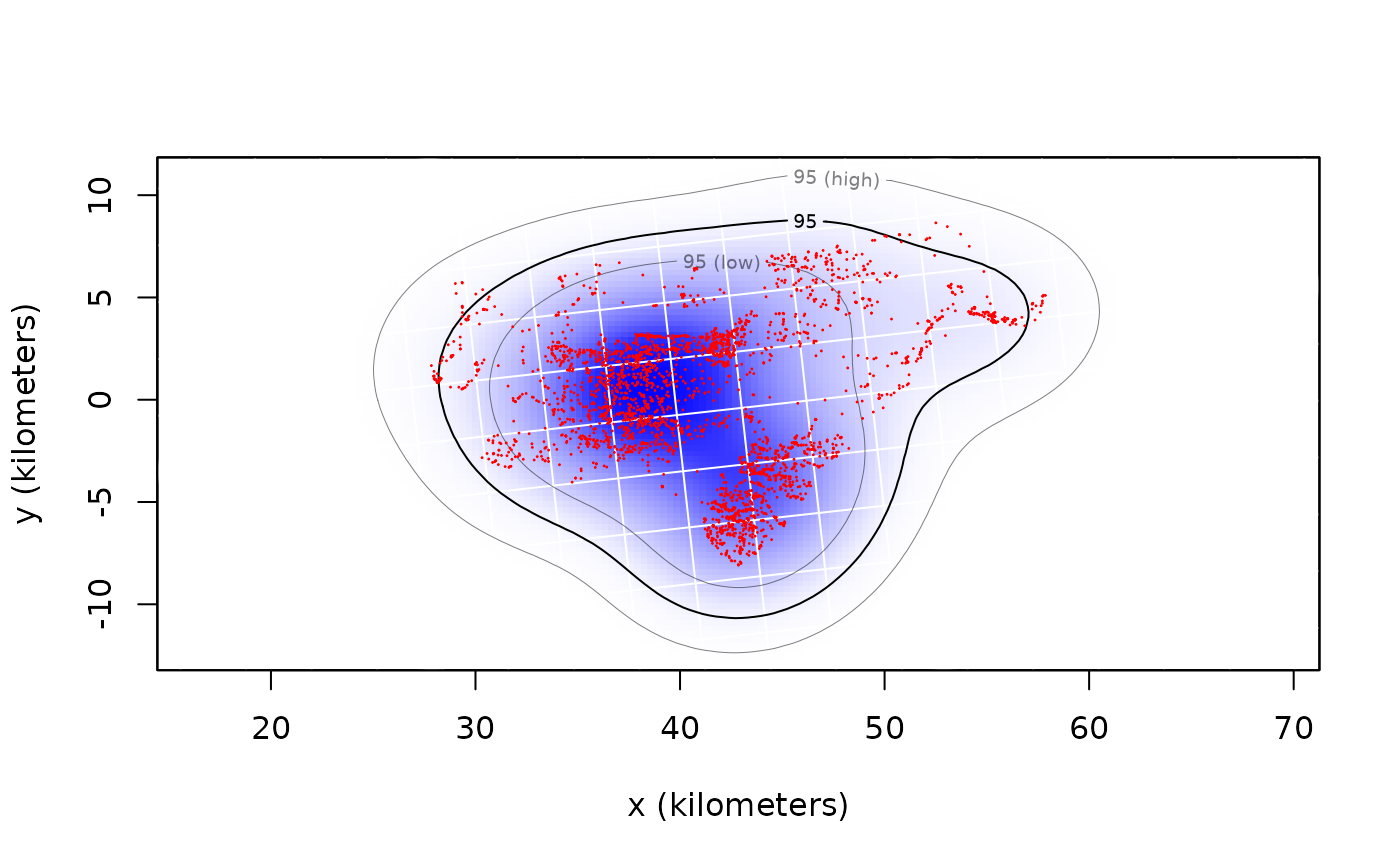
# Compute akde object
UD <- akde(DATA,FIT)
#> Default grid size of 3 minutes chosen for bandwidth(...,fast=TRUE).
# Plot data with AKDE
plot(DATA,UD=UD)
#> DOP values missing. Assuming DOP=1.
 # }
# }